We report the first isolation in Australia of a hypervirulent epidemic strain of Clostridium difficile, PCR ribotype 027. It was isolated from a 43-year-old woman with a permanent ileostomy, who appears to have been infected while travelling in the United States. The isolate was positive for toxin A, toxin B and binary toxin, and resistant to fluoroquinolone antimicrobials, and had characteristic deletions in the tcdC gene. All diagnostic laboratories and health care facilities in Australia should now be on high alert for this organism.
On her arrival back in Australia, the symptoms recurred. An ileoscopy on 4 February 2009 showed a single inflamed ulcerated area close to the stoma, and biopsy specimens of this area were reported as consistent with pseudomembranous enteritis and C. difficile infection (Box 1). Culture of the ileostomy fluid again resulted in the isolation of toxigenic C. difficile.
The isolate was determined to be positive for toxin A (tcdA), toxin B (tcdB), and binary toxin (CDT) by polymerase chain reaction (PCR) testing for toxin genes (tcdA, including the repetitive region, tcdB, and both the cdtA and cdtB binary toxin genes).1,2
penicillin, susceptible (S) (minimum inhibitory concentration [MIC], 0.75 mg/L);
clindamycin, S (MIC, 2 mg/L);
metronidazole, S (MIC, 0.38 mg/L);
levofloxacin, resistant (R) (MIC, > 32 mg/L);
moxifloxacin, R (MIC, 16 mg/L); and
vancomycin, S (MIC, 0.38 mg/L).
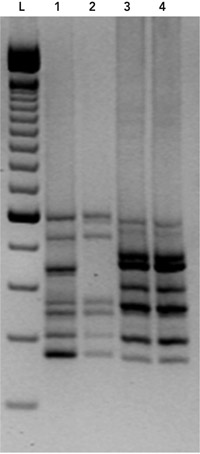
The tcdC gene (which encodes a negative regulator in toxin production) was sequenced and found to contain an 18-base-pair deletion, as well as a single nucleotide deletion at position 117,3 characteristic of the epidemic C. difficile strain, PCR ribotype 027. On the basis of these findings, the patient’s isolate was PCR ribotyped,4 which confirmed it to be C. difficile PCR ribotype 027 (Box 2). Because of the severity of the patient’s initial illness, she was treated with a further 14-day course of oral vancomycin (250 mg 6-hourly). Her condition improved, and she has remained well since, with two negative cultures for C. difficile since cessation of vancomycin.
A hypervirulent, epidemic strain of C. difficile, PCR ribotype 027, has been responsible for outbreaks of severe disease in North America and Europe. This organism is characterised by production of increased quantities of toxins A and B, plus an additional, binary toxin (actin-specific ADP-ribosyltransferase), and fluoroquinolone resistance. Overuse of fluoroquinolones is probably driving epidemic spread of this strain in North America and Europe, and attributable mortality in people aged over 60 years who are infected has been over 10%. There has been concern in Australia because of the lack of suitable surveillance systems to detect the entry of epidemic C. difficile into this country;5 this is the first report of such an occurrence.
This case is unusual because infection apparently occurred while the patient was travelling. Travel-associated C. difficile infection is extremely rare and, to our knowledge, acquisition of C. difficile during travel has never been proven. However, C. difficile is known to cause diarrhoea in travellers, as a result of antibiotics given either as prophylaxis before the journey, or as treatment for traveller’s diarrhoea afterwards. Indeed, in 1995, we reported three cases of laboratory-proven C. difficile infection following doxycycline administration for malaria prophylaxis.6 All three patients apparently acquired the organism outside Australia, although this could not be proven as no cultures for C. difficile were performed before travel. Six cases of C. difficile infection were recently reported in Spain in travellers who took antibiotics to treat an acute diarrhoeal episode and subsequently presented with prolonged or recurrent gastrointestinal symptoms, including diarrhoea.7 Interestingly, the first isolation of C. difficile PCR ribotype 027 in Austria was from a British tourist who was admitted to a hospital in Tyrol with a 5-day history of nausea, watery diarrhoea and lower abdominal pain. She was reportedly taking antibiotics prescribed by her physician to treat bronchitis, and the authors believed she acquired the strain in Great Britain before travel.8
We believe our patient most likely acquired C. difficile infection in New York City. According to the US Centers for Disease Control and Prevention, C. difficile PCR ribotype 027 has now been detected in 40 US states, including New York.9 It is less likely that she was infected while passing through Canada, even though PCR ribotype 027 is thought to be endemic in the western part of that country.10 In either case, as she had not been in contact with the health care system at that stage of the trip, community acquisition is most likely. Recent reports suggest that community acquisition of C. difficile is increasing worldwide.11
The lesions associated with C. difficile infection in humans are generally restricted to the colon, but pseudomembrane formation has been described in patients with an ileostomy.12 Our patient may have been at risk of C. difficile infection for two reasons. First, in ulcerative colitis, the intestine is known to be more readily colonised by C. difficile, and much of this colonisation occurs in the community rather than the health care setting.13 To what extent that risk is modified by colectomy is not known. The upper gastrointestinal tract microflora in patients with an ileostomy is similar to that of the colon 1 to 3 weeks after the ileostomy.14 Second, our patient had completed a course of antibiotics about 1 month earlier, and the “normal” microflora may not yet have re-established enough to provide any protection.
We were fortunate that this patient was seen at an institution that routinely cultures for C. difficile. Currently, most laboratories in Australia do not culture for C. difficile, instead relying on either enzyme immunoassays or PCR tests. As molecular typing is required to identify C. difficile PCR ribotype 027, at a minimum, all patients with severe, suspected C. difficile infection should have specimens cultured, and any isolates should be sent to a laboratory with expertise in identifying epidemic strains. General practioners and diagnostic laboratories need to be reminded that patients presenting with community-acquired or travel-related diarrhoea may have C. difficile infection. Periodic targeted surveillance with molecular typing of C. difficile isolates should be funded by government, as suggested previously,5 until more rapid molecular diagnostic tests to identify epidemic strains are developed.
2 PCR ribotyping of the Clostridium difficile strain isolated from the patient in February 2009
- 1. Kato H, Kato N, Watanabe K, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol 1998; 36: 2178-2182.
- 2. Stubbs S, Rupnik M, Gibert M, et al. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett 2000; 186: 307-312.
- 3. Curry SR, Marsh JW, Muto CA, et al. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol 2007; 45: 215-221.
- 4. Stubbs SL, Brazier JS, O’Neill GL, Duerden BI. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol 1999; 37: 461-463.
- 5. Riley TV. Epidemic Clostridium difficile. Med J Aust 2006; 185: 133-134. <MJA full text>
- 6. Golledge CL, Riley TV. Clostridium difficile-associated diarrhoea after doxycycline malaria prophylaxis. Lancet 1995; 345: 1377-1378.
- 7. Norman FF, Pérez-Molina J, Pérez de Ayala A, et al. Clostridium difficile-associated diarrhea after antibiotic treatment for traveler’s diarrhea. Clin Infect Dis 2008; 46: 1060-1063.
- 8. Indra A, Huhulescu S, Hasenberger P, et al. First isolation of Clostridium difficile PCR ribotype 027 in Austria. Euro Surveill 2006 Sep 14; 11 (9): E060914.3.
- 9. Centers for Disease Control and Prevention. States with BI/NAP1/027 strain of C. difficile (N=40), October, 2008. http://www.cdc.gov/ncidod/dhqp/pdf/State_Map_NAP1_10_2008.pdf (accessed Apr 2009).
- 10. MacCannell DR, Louie TJ, Gregson DB, et al. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from eastern and western Canada. J Clin Microbiol 2006; 44: 2147-2152.
- 11. Kuijper EJ, van Dissel JT. Spectrum of Clostridium difficile infections outside health care facilities. CMAJ 2008; 179: 747-748.
- 12. Bartlett JG. Clostridium difficile: clinical considerations. Rev Infect Dis 1990; 12 Suppl 2: S243-S251.
- 13. Clayton EM, Rea MC, Shanahan F, et al. The vexed relationship between Clostridium difficile and inflammatory bowel disease: an assessment of carriage in an outpatient setting among patients in remission. [Published online ahead of print, Am J Gastroenterol 2009; 24 Mar: doi: 10.1038/ajg.2009.4.]
- 14. Vince A, O’Grady F, Dawson AM. The development of ileostomy flora. J Infect Dis 1973; 128: 638-641.





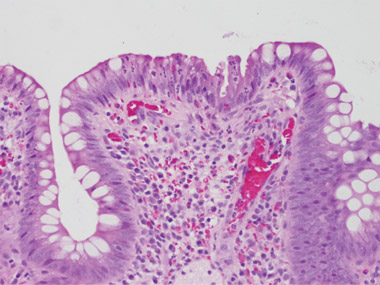
We are grateful to Professor Vincent Caruso (St John of God Hospital Pathology, Western Australia) for providing the photomicrographs.