The known: Testing people with colorectal cancer for Lynch syndrome has been found to be cost‐effective in some developed countries.
The new: In the first Australian cost‐effectiveness evaluation of systematic testing for Lynch syndrome in people with incident colorectal cancer that included all feasible combinations of relevant testing and triage options, the cost‐effectiveness ratios for universal tumour testing strategies for identifying mismatch repair deficiency (dMMR), compared with not testing, were similar. Universal gene panel testing was not cost‐effective compared with universal tumour testing strategies.
The implications: Our analysis supports routine dMMR tumour testing of people with incident colorectal cancer for guiding genetic testing for Lynch syndrome.
Lynch syndrome (LS), or hereditary non‐polyposis colorectal cancer, is an autosomal dominant cancer susceptibility disorder caused by constitutional mutations in four DNA mismatch repair (MMR) genes, MLH1, MSH2, MSH6, and PMS2.1 LS is associated with increased risk of developing a range of cancers, particularly colorectal cancer (CRC).1 Patients with CRC tumours exhibiting MMR deficiency (dMMR) but not the somatic BRAF V600E mutation or hypermethylation of the MLH1 promoter can be referred for germline genetic tests for LS.2 If LS is confirmed, cascade germline genetic testing can be made available to at‐risk relatives, and a range of cancer risk management options, including colonoscopic surveillance and prophylactic surgery, can be offered to reduce the morbidity and mortality associated with the syndrome.3
In some countries, including the United Kingdom,4 routine testing for LS is recommended for patients with incident CRC. In Australia, there is no national LS testing policy and the availability and practice of testing varies between clinicians, pathology providers, and states and territories.5 The most recent clinical CRC management practice guidelines encourage dMMR tumour testing for all patients with CRC as a “practice point” (a recommendation based on expert opinion and formulated in a consensus process).6 The Royal College of Pathologists of Australasia recently submitted an application to the Medical Services Advisory Committee requesting funding of germline gene panel testing for heritable mutations associated with increased risk of CRC and endometrial cancer; the panel of 11 genes includes the four MMR genes and the epithelial cell adhesion molecule gene (EPCAM).7 Two recent economic evaluations in Australia found that routine LS testing of people with incident CRC can be cost‐effective.7,8 However, no published assessment of systematic LS testing in Australia has taken into account all feasible combinations of relevant testing and triage options, including those for somatic mutation testing after dMMR immunohistochemistry or microsatellite instability testing.
A comprehensive evaluation of all relevant testing and triage options, including all benefits to probands and family members as well as the downstream cost benefits related to cancer prevention, would be appropriate. We therefore evaluated the health impact and cost‐effectiveness of systematic testing of people with incident CRC for LS, with the aim of providing evidence that could inform a national LS testing policy. Our specific aims were to determine the most cost‐effective LS testing strategy for people with incident CRC, and to estimate the health and economic impacts of limiting testing to specific CRC diagnosis age ranges and of different colonoscopic surveillance intervals for confirmed LS carriers.
Methods
We investigated the impact of various LS testing strategies in a micro‐simulation model (Policy1–Lynch; Supporting Information, section 1). We explicitly modelled the cost of testing all patients diagnosed with CRC during 2017, with detailed modelling of outcomes for patients identified as LS carriers (probands) and their at‐risk relatives throughout their lifetimes (censored at 100 years) (Box 1, Box 2; for detailed clinical management pathways: Supporting Information, section 2). For confirmed cases of LS (probands and relatives), we modelled ongoing colonoscopic surveillance. The detailed assumptions and parameter values are provided in the online Supporting Information, sections 3–8.
Analysis
We performed the analysis in three stages. In Stage 1 (baseline), eight testing strategies were examined (Supporting Information, section 2): no testing (as comparator, strategy 1); universal dMMR tumour testing (immunohistochemistry or microsatellite instability testing) with or without somatic BRAF V600E or MLH1 promoter methylation testing, followed by germline gene panel testing for confirmation of LS (strategies 2–7); and universal germline gene panel testing (strategy 8). We assumed that all patients diagnosed with CRC in 2017 would be tested for LS (ie, no age limit) and that confirmed LS carriers (probands and relatives) undertake annual colonoscopic surveillance until age 70.9
In Stage 2, we further investigated the impact of key parameters in an exploratory analysis of both the most cost‐effective strategy in Stage 1 and the universal germline gene panel testing strategy. We specifically modelled the effects on cost‐effectiveness of the CRC diagnosis ceiling age for LS testing (cancers diagnosed before 50, 60 or 70 years of age, or no age limit), and of the colonoscopic surveillance interval (one or two years). We also performed a supplementary analysis in which the colonoscopic surveillance adherence rate was reduced from 80% to 70% for all Stage 2 strategies.
In Stage 3, we performed a series of one‐way sensitivity analyses to investigate the effects of key parameters on the cost‐effectiveness of the most cost‐effective testing strategy identified in Stage 1 and the universal gene panel testing strategy.
Assumptions regarding the natural history of colorectal cancer in patients with Lynch syndrome and the effect of colonoscopic surveillance
We adopted a similar approach to that applied by the UK Health Technology Assessment report, using published parameters for developing our natural history model.10,11 Briefly, CRC development in LS carriers was modelled as cumulative CRC risk, with and without colonoscopic surveillance, for incident CRC and for second CRC in treated individuals.12,13,14 CRC incidence rates for people without LS in 2017 were based on 2014 Australian population‐based sex‐ and age‐specific CRC incidence (the most recent available data at the time of analysis).15 We assumed that an individual could develop up to two CRCs during their lifetime (ie, up to one metachronous CRC).
In Stage 1 and 2 analyses, we assumed that colonoscopic surveillance (with polypectomy if required) reduces the incidence of CRC and also downstages a proportion of the cancers not prevented. The estimated hazard ratio (HR) for first CRC in LS carriers undergoing 2‐ or 3‐yearly colonoscopic surveillance (v no surveillance) was 0.387,10,11,12 for those undergoing annual surveillance it was assumed to be 0.3. We analysed stage‐specific CRC 5‐year survival for people with CRC.16,17 Overall population life tables were used for calculating mortality from other causes (Supporting Information, section 4).
Assumptions regarding diagnostic test accuracy and referral adherence
Pooled estimates of sensitivity and specificity for MMR immunohistochemistry and microsatellite instability testing were used.11 The sensitivity and specificity of gene panel testing were each assumed to be 100%.18 We did not explicitly model colonoscopy test characteristics, as the reduction in CRC incidence associated with regular colonoscopic surveillance already captured the sensitivity and specificity of colonoscopy as part of the overall effectiveness of surveillance (Supporting Information, section 5). We also assumed all patients with CRC are tested for dMMR in strategies 2–7 and 90% consent to gene panel testing in strategies 2–8. We assumed that 78% of relatives attended genetic counselling,19 and that 77% of those attending consented to predictive genetic testing.19 We also assumed that the initial uptake of colonoscopic surveillance by probands and relatives was 90% and that the annual adherence rate was 80% (Supporting Information, section 6).
Assumptions regarding the family composition model
We assumed that each LS proband has a mean of six relatives eligible for cascade testing (equal numbers of children, siblings, and siblings' children), of whom 1.42 are found to be LS carriers after predictive genetic testing. This number was estimated by multiplying the number of eligible relatives (six) by the proband's referral rate for relatives (0.90) and the relatives' adherence rates to genetic counselling (0.78) and predictive genetic testing (0.77), and by the expected proportion of relatives with LS (44%) (Supporting Information, section 7).11
Assumptions regarding costs, utilities, and health economic parameters
We conducted the analysis from the perspective of the health care provider (Medicare), and included costs (2017 prices in Australian dollars) for testing, diagnosis, surveillance, and treatment procedures (Supporting Information, section 8). A discount rate of 5% was applied to both costs and health outcomes, and we assumed that a strategy was cost‐effective compared with no testing if the indicative willingness‐to‐pay was lower than $30 000–$50 000 per life‐year saved (LYS).20,21 We did not calculate quality‐adjusted life‐years (QALYs) because the available data were insufficient for informing utility weights for testing, diagnostic confirmation, predictive testing of relatives, and subsequent sequelae for surveillance. An incremental cost‐effectiveness ratio (ICER) was calculated in Stage 1 to determine which strategy was most cost‐effective.
Estimation of cost‐effectiveness compared with no testing strategy
We explicitly modelled the cost of testing all patients diagnosed with CRC in 2017, based on 2014 Australian population‐based sex‐ and age‐specific CRC incidence (the most recent available data)14 and the prevalence of LS carriers among patients with incident CRC. We simulated the lifetimes of one million patients with CRC and LS in each 5‐year age group and their at‐risk relatives, assuming the mean number of at‐risk relatives confirmed to have LS followed by predictive genetic testing to be 1.42 per proband. Aggregated health and resource outcomes for LS testing strategies for the cohort of LS carriers identified in 2017 are reported as numbers per 2420 LS carriers (1000 patients with CRC with LS and their 1420 relatives with confirmed LS) over their lifetimes (to 100 years).
Ethics approval
Ethics approval was not required for our modelled simulation based on publicly available aggregated data.
Results
In the Stage 1 (baseline) analysis, the cost‐effectiveness of strategies involving dMMR testing of CRC patients (strategies 2–7), compared with no testing, ranged between $28 915/LYS (strategy 3) and $31 904/LYS (strategy 5) (Box 3; Box 4, A). The most cost‐effective strategy — immunohistochemistry and reflex BRAF V600E testing followed by gene panel testing to confirm LS (strategy 3) — would require an additional 30 995 colonoscopies over the lifetimes of 2420 LS carriers and would avert 189 CRC deaths (164 extra colonoscopies to avert one CRC death) (Box 5). The discounted costs for immunohistochemistry with BRAF V600E testing are $10 645 per LS proband identified and $7044 including both proband and identified LS‐positive relatives (detailed results not shown). Compared with immunohistochemistry, universal germline gene panel testing would avert a further three deaths over the lifetimes of 2420 LS carriers, but its ICER ($2 411 933/LYS; Box 3) means that this strategy would not be cost‐effective compared with the dMMR tumour testing strategies.
In the Stage 2 analysis, the most cost‐effective testing strategy was MMR immunohistochemistry and reflex BRAF V600E testing, with 2‐yearly colonoscopic surveillance of confirmed LS carriers. The estimated ICERs were $11 525/LYS (age limit, < 50 years), $11 711/LYS (age limit, < 60 years), $12 106/LYS (age limit, < 70 years), and $32 153/LYS (no age limit), with more life‐years saved at increased cost as the age cut‐off is eased (Box 3; Box 4, B). An additional 4778–15 860 colonoscopies would be required over the lifetimes of 2420 LS carriers, and 46–181 CRC deaths would be averted (88–103 extra colonoscopies per averted CRC death) (Box 5). Universal germline gene panel testing with 2‐yearly surveillance would avert up to three more CRC deaths, but would not be as cost‐effective as universal dMMR tumour testing strategies. The ICER for universal gene panel testing strategy without age limit and annual colonoscopic surveillance (strategy 8) was $1 951 947/LYS. Reducing the colonoscopic surveillance adherence rate to 70% slightly reduced the cost‐effectiveness of these testing strategies, but the ranking of strategies by cost‐effectiveness remained the same; the ICER for immunohistochemistry/BRAF V600E testing ranged between $12 722/LYS and $37 858/LYS (Supporting Information, section 9).
The major components of the undiscounted costs over the lifetimes of 2420 LS carriers associated with universal dMMR tumour testing in the Stage 1 (baseline) analysis were those for newly detected cancer treatment (range, $81 672 800–$82 555 200) and colonoscopic surveillance (range, $56 452 200–$57 350 400), with lower cost ranges for tumour testing ($2 815 080–$5 038 780), proband gene panel testing ($2 234 670–$6 353 170) and predictive targeted gene testing for relatives ($2 163 910–$2 165 240). In the Stage 2 analysis, colonoscopic surveillance costs over the lifetimes of 2420 LS carriers, with the same testing age cut‐off, were lower with 2‐yearly than with annual colonoscopic surveillance ($8 815 020–$29 261 400) (Box 6).
For a given testing strategy and age limit, the number of life‐years saved by annual colonoscopic surveillance was not substantially greater than with 2‐yearly colonoscopic surveillance, but the costs were substantially higher, and the cost difference increased with rising age cut‐off. Testing strategies including annual colonoscopic surveillance were consequently less cost‐effective than those with 2‐yearly surveillance (Box 4, B).
In the Stage 3 sensitivity analyses, the parameter with the greatest influence on cost‐effectiveness was the assumed impact of colonoscopic surveillance on CRC incidence; that is, colonoscopic surveillance down‐stages diagnosed cancers but does not reduce the incidence (Box 7).
Discussion
We have reported one of the most comprehensive evaluations undertaken in any country of the cost‐effectiveness of systematic testing, cascade testing, and subsequent surveillance for LS. We found that all universal dMMR tumour testing strategies for people with incident CRC, without age limit and with annual colonoscopic surveillance of confirmed LS carriers, were similarly cost‐effective (compared with no testing) at an indicative willingness‐to‐pay threshold of $30 000–$50 000/LYS; the most cost‐effective strategy was immunohistochemistry with reflex BRAF V600E testing. Universal dMMR tumour testing strategies could reduce the number of CRC deaths by 184–189 while increasing the number of colonoscopies by 30 597–31 084 over the lifetimes of 1000 people with CRC and LS and 1420 relatives confirmed to be LS carriers (164–166 additional colonoscopies per death averted).
Our assumption that a mean six relatives per proband would be eligible for cascade genetic testing was probably conservative, and the benefits of systematic LS testing could therefore be greater than estimated. Life expectancy in Australia is comparatively high, which could also increase the cost‐effectiveness of interventions, such as LS testing, that chiefly extend the longevity of older people. Based on the number of people diagnosed with incident CRC in Australia (about 15 000 in 201414) and the estimated proportion of LS carriers (2.8%),22 about 420 probands could be identified per year, or 1000 probands over about 2.5 years. Routine dMMR tumour testing could therefore avert as many as 80 CRC deaths per year, and would require 13 000 extra colonoscopies per year, in addition to the 100 000–125 000 colonoscopies generated by referrals and surveillance under the National Bowel Cancer Screening Program.16
We found that 2‐yearly colonoscopic surveillance was more cost‐effective than annual surveillance, while assuming that CRC incidence is reduced a further 10% by annual surveillance compared with 2‐yearly surveillance. Recent studies found no statistically significant differences by surveillance interval in cumulative CRC incidence or CRC stage at diagnosis in people with LS.23,24 Ongoing quality colonoscopy is required to maximise the benefits of any strategy. We also found that universal germline gene panel testing is not as cost‐effective as dMMR test strategies, but its cost could decline substantially in the near future. Were universal gene panel testing introduced, it is likely that dMMR tumour testing would still be employed in clinical settings, both to guide treatment decisions for a subgroup of patients with dMMR tumours and to assist interpretation of genetic test results (eg, variants of uncertain significance). If further evidence for the role of dMMR testing in guiding cancer treatment accrues, the cost‐effectiveness of routine testing might require re‐assessment.
Several factors determine which laboratory test is chosen for identifying dMMR and excluding somatic MMR mutations. Immunohistochemistry is more convenient because it can be semi‐automated and performed in any histopathology laboratory, whereas microsatellite instability testing, which is more complicated, is available in fewer laboratories.25BRAF V600E reflex testing for dMMR in tumour samples is appropriate only for samples from patients with CRC. MLH1 methylation testing is necessary when testing for LS in people with endometrial or other LS‐related cancers; as MLH1 methylation testing is more complex, only some laboratories can perform it. In practice, the choice of universal dMMR and triage testing strategies will depend on local test availability and laboratory‐specific considerations.
Limitations
We focused on life‐years saved rather than quality‐adjusted life‐years because comprehensive data on health state preferences (ie, utilities) needed to calculate quality‐adjusted life‐years for people at risk of LS are not available. Although we assumed that gene panel testing is 100% sensitive (to simplify modelling), its reported sensitivity is 99.4%;18 that is, a very small proportion of LS carriers will not be detected. Further, we did not consider the impact of an incidental diagnosis (eg, non‐LS hereditary cancers) following universal germline gene panel testing. As in earlier key analyses in the UK,10,11 we did not consider the possibility of systematic LS testing in the context of cancers at other sites (eg, endometrial cancer), nor did we consider the possible health benefits or likely harms and costs of gynaecological surveillance (eg, transvaginal ultrasound, aspiration biopsy) or prophylactic surgery (eg, total abdominal hysterectomy with or without salpingo‐oophorectomy) for women identified as LS carriers by systematic testing of people with CRC. However, there is no conclusive evidence for the effectiveness and health benefits of gynaecological surveillance. While we considered neither the health benefits of testing non‐MMR genes nor endometrial cancer outcomes, we fully captured all downstream procedures, costs, and benefits involved in implementing a routine LS testing program for people with CRC. Our analysis accordingly complements the analysis of 11‐gene panel testing currently underway.7
Conclusion
At an indicative willingness‐to‐pay threshold of $30 000–$50 000/life‐year saved, a range of universal dMMR tumour testing strategies for systematic LS testing of people with incident CRC are likely to be similarly cost‐effective in Australia. Universal gene panel testing is not yet cost‐effective, but should be re‐evaluated should its costs drop, as is expected.
Box 1 – Testing strategies to identify Lynch syndrome mutation carriers among people diagnosed with incident colorectal cancer (probands)
- Strategy 1. No testing (comparator).
- Strategy 2. DNA mismatch repair (dMMR) immunohistochemistry (IHC) four‐panel test → diagnostic germline gene panel testing if IHC result is abnormal (absence of staining [= loss of expression] for MLH1/MSH2/MSH6/PMS2 proteins).
- Strategy 3. dMMR IHC four‐panel test → somatic BRAF V600E testing if IHC result for MLH1 is abnormal → diagnostic germline gene panel testing if somatic BRAF V600E test result is negative.
- Strategy 4. dMMR IHC four‐panel test → somatic MLH1 promoter hypermethylation testing if IHC result for MLH1 is abnormal → diagnostic germline gene panel testing if somatic methylation test result is negative.
- Strategy 5. Molecular dMMR microsatellite instability (MSI) test → diagnostic germline gene panel testing if MSI result is abnormal (ie, high instability = MSI‐H).
- Strategy 6. Molecular dMMR MSI test → somatic BRAF V600E testing if MSI result is abnormal → diagnostic germline gene panel testing if somatic BRAF V600E test result is negative.
- Strategy 7. Molecular dMMR MSI test → somatic MLH1 promoter hypermethylation testing if MSI result is abnormal → diagnostic germline gene panel testing if somatic methylation test result is negative.
- Strategy 8. Universal germline gene panel testing in all CRC cases.
- Probands undertake annual colonoscopic surveillance from colorectal diagnosis until age 70 years
- Relatives undertake annual colonoscopic surveillance from age 25 (or from the age when Lynch syndrome was confirmed) until age 70 years
Box 2 – Flow chart of testing and management strategies for Lynch syndrome (LS) probands and relatives

Box 3 – Health economic outcomes associated with testing people with incident colorectal cancer (CRC) diagnosed in Australia during 2017 for Lynch syndrome (LS), compared with no testing (per 1000 people with incident CRC with LS and 1420 relatives with confirmed LS)
|
Testing strategy (age range for testing; colonoscopic surveillance interval) |
Discounted costs |
Discounted life‐years saved (LYS) |
Cost‐effectiveness ratio: testing v no testing |
Incremental cost‐effectiveness ratio (ICER) |
|||||||||||
|
|
|||||||||||||||
|
Stage 1 analysis |
|
|
|
|
|||||||||||
|
1. No testing |
$12 640 |
14.0917 |
— |
— |
|||||||||||
|
7. MSI + MLH1 methylation (no age limit; 1 year) |
$19 145 |
14.3061 |
$30 338/LYS |
Dominated |
|||||||||||
|
6. MSI + BRAF V600E (no age limit; 1 year) |
$19 130 |
14.3079 |
$30 020/LYS |
Dominated |
|||||||||||
|
5. MSI (no age limit; 1 year) |
$19 646 |
14.3113 |
$31 904/LYS |
Dominated |
|||||||||||
|
4. IHC + MLH1 methylation (no age limit; 1 year) |
$19 075 |
14.3121 |
$29 196/LYS |
Dominated |
|||||||||||
|
3. IHC + BRAF V600E (no age limit; 1 year) |
$19 068 |
14.3140 |
$28 915/LYS |
$28 915/LYS |
|||||||||||
|
2. IHC (no age limit; 1 year) |
$19 249 |
14.3148 |
$29 625/LYS |
$227 000/LYS |
|||||||||||
|
8. Universal gene panel testing (no age limit; 1 year) |
$26 485 |
14.3178 |
$61 235/LYS |
$2 411 933/LYS |
|||||||||||
|
Stage 2 analysis |
|
|
|
|
|||||||||||
|
1. No testing |
$12 640 |
14.0917 |
— |
— |
|||||||||||
|
3.3. IHC + BRAF V600E (< 50 years; 2 years) |
$13 244 |
14.1441 |
$11 525/LYS |
$11 525/LYS |
|||||||||||
|
3.2. IHC + BRAF V600E (< 50 years; 1 year) |
$14 056 |
14.1451 |
$26 507/LYS |
Dominated |
|||||||||||
|
8.3. Universal gene panel testing (< 50 years; 2 years) |
$13 746 |
14.1453 |
$20 632/LYS |
Dominated |
|||||||||||
|
8.2. Universal gene panel testing (< 50 years; 1 year) |
$14 566 |
14.1461 |
$35 397/LYS |
Dominated |
|||||||||||
|
3.5. IHC + BRAF V600E (< 60 years; 2 years) |
$14 155 |
14.2219 |
$11 636/LYS |
$11 711/LYS |
|||||||||||
|
8.5. Universal gene panel testing (< 60 years; 2 years) |
$15 989 |
14.2246 |
$25 199/LYS |
Dominated |
|||||||||||
|
3.4. IHC + BRAF V600E (< 60 years; 1 year) |
$16 261 |
14.2263 |
$26 898/LYS |
Dominated |
|||||||||||
|
8.4. Universal gene panel testing (< 60 years; 1 year) |
$18 131 |
14.2290 |
$39 992/LYS |
Dominated |
|||||||||||
|
3.7. IHC + BRAF V600E (< 70 years; 2 years) |
$14 832 |
14.2778 |
$11 777/LYS |
$12 106/LYS |
|||||||||||
|
8.7. Universal gene panel testing (< 70 years; 2 years) |
$18 404 |
14.2815 |
$30 369/LYS |
Dominated |
|||||||||||
|
3.6. IHC + BRAF V600E (< 70 years; 1 year) |
$17 803 |
14.2845 |
$26 776/LYS |
Dominated |
|||||||||||
|
8.6. Universal gene panel testing (< 70 years; 1 year) |
$21 424 |
14.2883 |
$44 681/LYS |
Dominated |
|||||||||||
|
3.1. IHC + BRAF V600E (no age limit; 2 years) |
$15 735 |
14.3059 |
$14 450/LYS |
$32 153/LYS |
|||||||||||
|
8.1. Universal gene panel testing (no age limit; 2 years) |
$23 101 |
14.3101 |
$47 899/LYS |
Dominated |
|||||||||||
|
3. IHC + BRAF V600E (no age limit; 1 year) |
$19 068 |
14.3140 |
$28 915/LYS |
$411 432/LYS |
|||||||||||
|
8. Universal gene panel testing (no age limit; 1 year) |
$26 485 |
14.3178 |
$61 235/LYS |
$1 951 947/LYS |
|||||||||||
|
|
|||||||||||||||
|
IHC = immunohistochemistry; MSI = microsatellite instability. * Costs and life‐years are each discounted by 5%. † Difference in costs divided by difference in LYS for strategy v no testing. ‡ Relative to the next most cost‐effective strategy. “Dominated” indicates that a strategy has either higher costs or a higher cost per LYS than a more effective strategy. |
|||||||||||||||
Box 4 – Discounted costs and numbers of life‐years saved associated with testing people with incident colorectal cancer (CRC) diagnosed in Australia during 2017 for Lynch syndrome (LS)
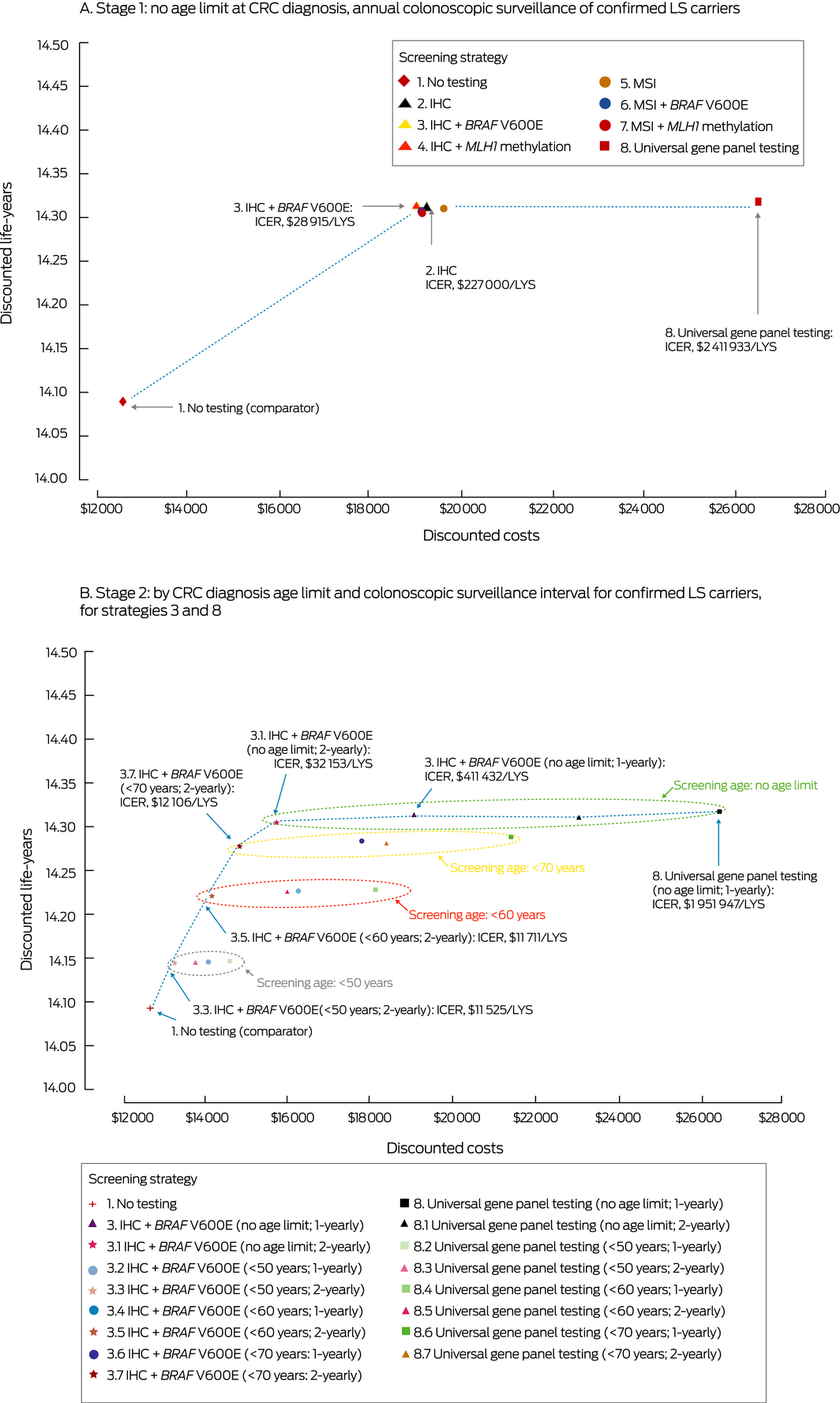
IHC = immunohistochemistry; MSI = microsatellite instability; ICER = incremental cost‐effectiveness ratio; LYS = life‐year saved. Strategies not on the dotted line are dominated; that is, thy have either higher costs or a higher cost per LYS than a more effective strategy.
Box 5 – Health and resource outcomes associated with testing people with incident colorectal cancer (CRC) diagnosed in Australia during 2017 for Lynch syndrome (LS), compared with no testing (per 1000 people with incident CRC with LS and 1420 relatives with confirmed LS)
|
Testing strategy (age range for testing; colonoscopic surveillance interval) |
Cases of cancer |
Cancer deaths |
Colonoscopies |
Cancer deaths averted |
Colonoscopies to avert one death |
||||||||||
|
|
|||||||||||||||
|
Stage 1 analysis |
|
|
|
|
|
||||||||||
|
1. No testing |
1566 |
630 |
— |
— |
— |
||||||||||
|
7. MSI + MLH1 methylation (no age limit; 1 year) |
1248 |
446 |
30 597 |
184 |
166 |
||||||||||
|
6. MSI + BRAF V600E (no age limit; 1 year) |
1246 |
445 |
30 697 |
185 |
166 |
||||||||||
|
5. MSI (no age limit; 1 year) |
1243 |
442 |
30 865 |
187 |
165 |
||||||||||
|
4. IHC + MLH1 methylation (no age limit; 1 year) |
1243 |
442 |
30 952 |
188 |
165 |
||||||||||
|
3. IHC + BRAF V600E (no age limit; 1 year) |
1242 |
441 |
30 995 |
189 |
164 |
||||||||||
|
2. IHC (no age limit; 1 year) |
1241 |
440 |
31 084 |
189 |
164 |
||||||||||
|
8. Universal gene panel testing (no age limit; 1 year) |
1239 |
438 |
31 257 |
192 |
163 |
||||||||||
|
Stage 2 analysis |
|
|
|
|
|
||||||||||
|
1. No testing |
1566 |
630 |
— |
— |
— |
||||||||||
|
3.3. IHC + BRAF V600E (< 50 years; 2 years) |
1500 |
583 |
4778 |
46 |
103 |
||||||||||
|
3.2. IHC + BRAF V600E (< 50 years; 1 year) |
1491 |
582 |
9457 |
48 |
198 |
||||||||||
|
8.3. Universal gene panel testing (< 50 years; 2 years) |
1500 |
583 |
4837 |
47 |
103 |
||||||||||
|
8.2. Universal gene panel testing (< 50 years; 1 year) |
1491 |
581 |
9571 |
49 |
196 |
||||||||||
|
3.5. IHC + BRAF V600E (< 60 years; 2 years) |
1398 |
518 |
10 642 |
112 |
95 |
||||||||||
|
8.5. Universal gene panel testing (< 60 years; 2 years) |
1396 |
516 |
10 756 |
114 |
94 |
||||||||||
|
3.4. IHC + BRAF V600E (< 60 years; 1 year) |
1375 |
513 |
20 976 |
117 |
180 |
||||||||||
|
8.4. Universal gene panel testing (< 60 years; 1 year) |
1374 |
511 |
21 197 |
119 |
179 |
||||||||||
|
3.7. IHC + BRAF V600E (< 70 years; 2 years) |
1319 |
471 |
14 251 |
159 |
90 |
||||||||||
|
8.7. Universal gene panel testing (< 70 years; 2 years) |
1317 |
468 |
14 383 |
161 |
89 |
||||||||||
|
3.6. IHC + BRAF V600E (< 70 years; 1 year) |
1287 |
464 |
27 970 |
165 |
169 |
||||||||||
|
8.6. Universal gene panel testing (< 70 years; 1 year) |
1284 |
462 |
28 226 |
168 |
168 |
||||||||||
|
3.1. IHC + BRAF V600E (no age limit; 2 years) |
1279 |
449 |
15 860 |
181 |
88 |
||||||||||
|
8.1. Universal gene panel testing (no age limit; 2 years) |
1275 |
446 |
15 999 |
184 |
87 |
||||||||||
|
3. IHC + BRAF V600E (no age limit; 1 year) |
1242 |
441 |
30 995 |
189 |
164 |
||||||||||
|
8. Universal gene panel testing (no age limit; 1 year) |
1239 |
438 |
31 257 |
192 |
163 |
||||||||||
|
|
|||||||||||||||
|
IHC = immunohistochemistry; MSI = microsatellite instability. |
|||||||||||||||
Box 6 – Lifetime discounted costs associated with testing people with incident colorectal cancer (CRC) diagnosed in Australia during 2017 for Lynch syndrome (LS), compared with no testing (per 1000 people with incident CRC with LS and 1420 relatives with confirmed LS)
|
Testing strategy (age range for testing; colonoscopic surveillance interval) |
Cost (proportion of total cost) |
||||||||||||||
|
Total costs |
Proband gene panel testing |
Relatives' genetic testing and counselling |
dMMR tumour testing |
Cancer treatment |
Colonoscopy |
||||||||||
|
|
|||||||||||||||
|
Stage 1 analysis |
|
|
|
|
|
|
|||||||||
|
1. No testing |
$110 474 000 |
— |
— |
— |
$110 474 000 (100%) |
— |
|||||||||
|
7. MSI + MLH1 methylation (no age limit; 1 year) |
$148 479 000 |
$2 268 750 (2%) |
$2 163 910 (1%) |
$5 038 780 (3%) |
$82 555 200 (56%) |
$56 452 200 (38%) |
|||||||||
|
6. MSI + BRAF V600E (no age limit; 1 year) |
$148 412 000 |
$2 234 670 (2%) |
$2 165 240 (1%) |
$5 038 730 (3%) |
$82 338 100 (55%) |
$56 635 400 (38%) |
|||||||||
|
5. MSI (no age limit; 1 year) |
$150 300 000 |
$6 353 170 (4%) |
$2 164 630 (1%) |
$2 815 080 (2%) |
$82 021 700 (55%) |
$56 945 300 (38%) |
|||||||||
|
4. IHC + MLH1 methylation (no age limit; 1 year) |
$148 225 000 |
$3 395 100 (2%) |
$2 164 830 (1%) |
$3 665 380 (2%) |
$81 892 700 (55%) |
$57 106 900 (39%) |
|||||||||
|
3. IHC + BRAF V600E (no age limit; 1 year) |
$148 193 000 |
$3 386 450 (2%) |
$2 164 670 (1%) |
$3 665 260 (2%) |
$81 789 600 (55%) |
$57 186 600 (39%) |
|||||||||
|
2. IHC (no age limit; 1 year) |
$148 861 000 |
$4 858 440 (3%) |
$2 164 240 (1%) |
$2 815 080 (2%) |
$81 672 800 (55%) |
$57 350 400 (39%) |
|||||||||
|
8. Universal gene panel testing (no age limit; 1 year) |
$175 199 000 |
$34 007 500 (19%) |
$2 164 360 (1%) |
— |
$81 357 600 (46%) |
$57 669 300 (33%) |
|||||||||
|
Stage 2 analysis |
|
|
|
|
|
|
|||||||||
|
1. No testing |
$110 474 000 |
— |
— |
— |
$110 474 000 (100%) |
— |
|||||||||
|
3.3. IHC + BRAF V600E (< 50 years; 2 years) |
$114 170 000 |
$397 177 (< 1%) |
$459 442 (< 1%) |
$529 723 (< 1%) |
$103 969 000 (91%) |
$8 815 020 (8%) |
|||||||||
|
3.2. IHC + BRAF V600E (< 50 years; 1 year) |
$122 399 000 |
$397 207 (< 1%) |
$459 183 (< 1%) |
$529 606 (< 1%) |
$103 565 000 (85%) |
$17 448 200 (14%) |
|||||||||
|
8.3. Universal gene panel testing (< 50 years; 2 years) |
$116 060 000 |
$2 737 630 (2%) |
$459 216 (< 1%) |
— |
$103 939 000 (90%) |
$8 924 350 (8%) |
|||||||||
|
8.2. Universal gene panel testing (< 50 years; 1 year) |
$124 294 000 |
$2 737 530 (2%) |
$459 167 (< 1%) |
— |
$103 439 000 (83%) |
$17 658 000 (14%) |
|||||||||
|
3.5. IHC + BRAF V600E (< 60 years; 2 years) |
$117 501 000 |
$1 160 090 (1%) |
$1 176 740 (1%) |
$1 139 730 (1%) |
$94 389 500 (80%) |
$19 634 800 (17%) |
|||||||||
|
8.5. Universal gene panel testing (< 60 years; 2 years) |
$124 180 000 |
$9 014 050 (7%) |
$1 176 590 (1%) |
— |
$94 144 300 (76%) |
$19 844 700 (16%) |
|||||||||
|
3.4. IHC + BRAF V600E (< 60 years; 1 year) |
$135 409 000 |
$1 160 100 (1%) |
$1 176 150 (1%) |
$1 139 690 (1%) |
$93 231 700 (69%) |
$38 701 300 (29%) |
|||||||||
|
8.4. Universal gene panel testing (< 60 years; 1 year) |
$142 286 000 |
$9 013 810 (6%) |
$1 176 350 (1%) |
— |
$92 987 300 (65%) |
$39 108 900 (27%) |
|||||||||
|
3.7. IHC + BRAF V600E (< 70 years; 2 years) |
$119 303 000 |
$2 005 420 (2%) |
$1 791 260 (2%) |
$1 937 940 (2%) |
$87 275 300 (73%) |
$26 292 600 (22%) |
|||||||||
|
8.7. Universal gene panel testing (< 70 years; 2 years) |
$132 274 000 |
$17 064 300 (13%) |
$1 790 520 (1%) |
— |
$86 883 100 (66%) |
$26 536 500 (20%) |
|||||||||
|
3.6. IHC + BRAF V600E (< 70 years; 1 year) |
$142 963 000 |
$2 005 540 (1%) |
$1 790 300 (1%) |
$1 937 960 (1%) |
$85 625 000 (60%) |
$51 604 400 (36%) |
|||||||||
|
8.6. Universal gene panel testing (< 70 years; 1 year) |
$156 170 000 |
$17 064 100 (11%) |
$1 790 140 (1%) |
— |
$85 239 800 (55%) |
$52 076 300 (33%) |
|||||||||
|
3.1. IHC + BRAF V600E (no age limit; 2 years) |
$122 173 000 |
$3 386 260 (3%) |
$2 165 780 (2%) |
$3 665 230 (3%) |
$83 693 900 (69%) |
$29 261 400 (24%) |
|||||||||
|
8.1. Universal gene panel testing (no age limit; 2 years) |
$148 950 000 |
$34 007 900 (23%) |
$2 164 800 (1%) |
— |
$83 259 600 (56%) |
$29 517 400 (20%) |
|||||||||
|
3. IHC + BRAF V600E (no age limit; 1 year) |
$148 193 000 |
$3 386 450 (2%) |
$2 164 670 (1%) |
$3 665 260 (2%) |
$81 789 600 (55%) |
$57 186 600 (39%) |
|||||||||
|
8. Universal gene panel testing (no age limit; 1 year) |
$175 199 000 |
$34 007 500 (19%) |
$2 164 360 (1%) |
— |
$81 357 600 (46%) |
$57 669 300 (33%) |
|||||||||
|
|
|||||||||||||||
|
dMMR = DNA mismatch repair deficiency; IHC = immunohistochemistry; MSI = microsatellite instability. * Initial CRC treatment costs for the proband were not included in the analysis, as they were assumed to not be affected by LS testing. |
|||||||||||||||
Box 7 – One‐way sensitivity analyses of the cost‐effectiveness of Lynch syndrome (LS) testing (all ages, annual surveillance of confirmed LS carriers [probands and relatives]), compared with no testing*

CI = confidence interval; dMMR = mismatch repair deficiency; HR = hazard ratio; IHC = immunohistochemistry. * The change in cost‐effectiveness associated with the first indicated value for a parameter is depicted by the darker (left side) bars, which indicate lower cost‐effectiveness ratio (ie, more cost‐effective), the change associated with the second value is depicted by the lighter (right side) bars, which indicate higher cost‐effectiveness ratio (ie, less cost‐effective).Solid vertical line: Stage 1 cost‐effectiveness estimate; dotted vertical lines: indicative willingness‐to‐pay threshold $30 000–$50 000/life‐year saved.For sensitivity and specificity of dMMR tumour testing, 95% CIs of pooled estimates for IHC, microsatellite instability, BRAF V600E, and MLH1 hypermethylation tests were varied simultaneously.
Received 15 January 2019, accepted 1 June 2019
- Yoon‐Jung Kang1
- James Killen1
- Michael Caruana1
- Kate Simms1
- Natalie Taylor1
- Ian M Frayling2,3
- Tristan Snowsill4
- Nicola Huxley5
- Veerle MH Coupe6
- Suzanne Hughes1
- Victoria Freeman1
- Alex Boussioutas7,8
- Alison H Trainer9
- Robyn L Ward10,11
- Gillian Mitchell9
- Finlay A Macrae8
- Karen Canfell1,10,11
- 1 Cancer Research Division, Cancer Council New South Wales, Sydney, NSW
- 2 Institute of Medical Genetics, University Hospital of Wales, Cardiff, United Kingdom
- 3 Institute of Cancer and Genetics, Cardiff University, Cardiff, United Kingdom
- 4 University of Exeter Medical School, Exeter, United Kingdom
- 5 Centre for Health Economics, Monash Business School, Monash University, Melbourne, VIC
- 6 Amsterdam Public Health Research Institute, VU University Medical Center, Amsterdam, The Netherlands
- 7 University of Melbourne, Melbourne, VIC
- 8 Royal Melbourne Hospital, Melbourne, VIC
- 9 Parkville Familial Cancer Centre, Peter MacCallum Cancer Institute, Melbourne, VIC
- 10 University of Sydney, Sydney, NSW
- 11 University of New South Wales, Sydney, NSW
We acknowledge funding from the National Health and Medical Research Council (NHMRC; GNT1080246) and the Cancer Council NSW (STREP: SRP 13‐02). Karen Canfell receives salary support from the NHMRC (Career Development Fellowship APP1082989). Neither funding source had direct influence on study design or findings. We thank the investigators of the Inherited Cancer Connect Program for their advice on this study.
Karen Canfell is co‐Principal Investigator of an unrelated trial of cervical screening, funded by the Victorian Cytology Service (VCS), that has received equipment and funding from Roche Molecular Systems, which also manufactures assays for genetic testing that determine access to targeted therapies for colorectal cancer.
- 1. Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003; 348: 919–932.
- 2. Cancer Institute NSW. Genetic testing for heritable mutations in the mismatch repair genes (MMR‐genes). eviQ (Cancer Treatments Online); updated 29 Nov 2018. https://www.eviq.org.au/cancer-genetics/genetic-testing-for-heritable-mutations/619-genetic-testing-for-heritable-mutations-in-the (viewed July 2019).
- 3. Cancer Institute NSW. Referral guidelines for colorectal cancer or polyposis risk assessment and consideration of genetic testing. eviQ (Cancer Treatments Online); updated 24 Aug 2016. https://www.eviq.org.au/cancer-genetics/referral-guidelines/657-referral-guidelines-for-colorectal-cancer-or-p (viewed July 2019).
- 4. National Institute for Health and Care Excellence. Molecular testing strategies for Lynch syndrome in people with colorectal cancer (Diagnostic guidance, G27). Feb 2017. https://www.nice.org.uk/guidance/dg27 (viewed July 2019).
- 5. Mascarenhas L, Shanley S, Mitchell G, et al. Current mismatch repair deficiency tumor testing practices and capabilities: a survey of Australian pathology providers. Asia Pac J Clin Oncol 2018; 14: 417–425.
- 6. Cancer Council Australia Colorectal Cancer Guidelines Working Party. Clinical practice guidelines for the prevention, early detection and management of colorectal cancer. Revised 11 Sept 2018. https://wiki.cancer.org.au/australia/Guidelines:Colorectal_cancer (viewed July 2019).
- 7. Australian Department of Health, Medical Services Advisory Committee. Heritable mutations which increase risk in colorectal and endometrial cancer (Public Summary Document, Applications no. 1504). Updated 29 Nov 2018. http://msac.gov.au/internet/msac/publishing.nsf/Content/1504-public (viewed July 2019).
- 8. Cenin DR, Naber SK, Lansdorp‐Vogelaar I, et al. Costs and outcomes of Lynch syndrome screening in the Australian colorectal cancer population. J Gastroenterol Hepatol 2018; 33: 1737–1744.
- 9. Cancer Institute NSW. Risk management for Lynch syndrome. eviQ (Cancer Treatments Online); updated 17 June 2019. https://www.eviq.org.au/cancer-genetics/risk-management/1410-risk-management-for-lynch-syndrome#cancer-risk-management (viewed July 2019).
- 10. Snowsill T, Huxley N, Hoyle M, et al. A systematic review and economic evaluation of diagnostic strategies for Lynch syndrome. Health Technol Assess 2014; 18(58).
- 11. Snowsill T, Coelho H, Huxley N, et al. Molecular testing for Lynch syndrome in people with colorectal cancer: systematic reviews and economic evaluation. Health Technol Assess 2017; 21(51).
- 12. Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15‐year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000; 118: 829–834.
- 13. Bonadona V, Bonaïti B, Olschwang S, et al. French Cancer Genetics Network. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011; 305: 2304–2310.
- 14. Parry S, Win AK, Parry B, et al. Metachronous colorectal cancer risk for mismatch repair gene mutation carriers: the advantage of more extensive colon surgery. Gut 2011; 60: 950–957.
- 15. Australian Institute of Health and Welfare. Australian cancer incidence and mortality (ACIM) books: colorectal (bowel) cancer. Jan 2017. https://www.aihw.gov.au/getmedia/b928eae4-ec59-4aca-8324-c188809d9420/colorectal-bowel-cancer.xls.aspx (viewed July 2019).
- 16. Lew JB, St John DJB, Xu XM, et al. Long‐term evaluation of benefits, harms, and cost‐effectiveness of the National Bowel Cancer Screening Program in Australia: a modelling study. Lancet Public Health 2017; 2: e331–e340.
- 17. Lin KM, Shashidharan M, Ternent CA, et al. Colorectal and extracolonic cancer variations in MLH1/MSH2 hereditary nonpolyposis colorectal cancer kindreds and the general population. Dis Colon Rectum 1998; 41: 428–433.
- 18. Pritchard CC, Smith C, Salipante SJ, et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn 2012; 14: 357–366.
- 19. Barrow P. Hereditary colorectal cancer: registration, screening and prognostic biomarker analysis. Thesis: University of Manchester, 2015. https://www.research.manchester.ac.uk/portal/files/54567811/FULL_TEXT.PDF (viewed July 2019).
- 20. Australian Department of Health, the Pharmaceutical Benefits Scheme. Quadrivalent human papillomavirus (types 6, 11, 16, 18) recombinant vaccine, injection, 0.5 mL, Gardasil. Updated 2 Mar 2007. http://www.pbs.gov.au/info/industry/listing/elements/pbac-meetings/psd/2006-11/pbac-psd-gardasil-nov06 (viewed July 2019).
- 21. Lew JB, Simms KT, Smith MA, et al. National Cervical Screening Program Renewal: effectiveness modelling and economic evaluation in the Australian setting (assessment report) (MSAC Application No. 1276). Nov 2013. http://www.cancerscreening.gov.au/internet/screening/publishing.nsf/Content/E6A211A6FFC29E2CCA257CED007FB678/$File/Renewal%20Economic%20Evaluation.pdf (viewed July 2019).
- 22. Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 2008; 26: 5783–5788.
- 23. Engel C, Vasen HF, Seppälä T, et al. No difference in colorectal cancer incidence or stage at detection by colonoscopy among 3 countries with different Lynch syndrome surveillance policies. Gastroenterology 2018; 155: 1400–1409.e2.
- 24. Seppälä TT, Ahadova A, Dominguez‐Valentin M, et al. Lack of association between screening interval and cancer stage in Lynch syndrome may be accounted for by over‐diagnosis; a prospective Lynch syndrome database report. Hered Cancer Clin Pract 2019; 17: 8.
- 25. Wedden S, Miller K, Frayling IM, et al. Colorectal cancer stratification in the routine clinical pathway: a district general hospital experience. Appl Immunohistochem Mol Morphol 2019; 27: e54–e62.






Abstract
Objectives: To evaluate the health impact and cost‐effectiveness of systematic testing for Lynch syndrome (LS) in people with incident colorectal cancer (CRC) in Australia.
Design, setting, participants: We investigated the impact of LS testing strategies in a micro‐simulation model (Policy1–Lynch), explicitly modelling the cost of testing all patients diagnosed with incident CRC during 2017, with detailed modelling of outcomes for patients identified as LS carriers (probands) and their at‐risk relatives throughout their lifetimes. For people with confirmed LS, we modelled ongoing colonoscopic surveillance.
Main outcome measures: Cost‐effectiveness of six universal tumour testing strategies (testing for DNA mismatch repair deficiencies) and of universal germline gene panel testing of patients with incident CRC; impact on cost‐effectiveness of restricting testing by age at CRC diagnosis (all ages, under 50/60/70 years) and of colonoscopic surveillance interval (one, two years).
Results: The cost‐effectiveness ratio of universal tumour testing strategies (annual colonoscopic surveillance, no testing age limit) compared with no testing ranged from $28 915 to $31 904/life‐year saved (LYS) (indicative willingness‐to‐pay threshold: $30 000–$50 000/LYS). These strategies could avert 184–189 CRC deaths with an additional 30 597–31 084 colonoscopies over the lifetimes of 1000 patients with incident CRC with LS and 1420 confirmed LS carrier relatives (164–166 additional colonoscopies/death averted). The most cost‐effective strategy was immunohistochemistry and BRAF V600E testing (incremental cost‐effectiveness ratio [ICER], $28 915/LYS). Universal germline gene panel testing was not cost‐effective compared with universal tumour testing strategies (ICER, $2.4 million/LYS). Immunohistochemistry and BRAF V600E testing was cost‐effective at all age limits when paired with 2‐yearly colonoscopic surveillance (ICER, $11 525–$32 153/LYS), and required 4778–15 860 additional colonoscopies to avert 46–181 CRC deaths (88–103 additional colonoscopies/death averted).
Conclusions: Universal tumour testing strategies for guiding germline genetic testing of people with incident CRC for LS in Australia are likely to be cost‐effective compared with no testing. Universal germline gene panel testing would not currently be cost‐effective.